A Deep Dive Into Our Paper on Patient-Specific Biomolecular Instruction Tuning of Graph LLMs
Combining patient-specific protein networks with instruction-tuned LLMs results in a system that truly understands the biological story behind each individual patient's data
Two weeks ago, Standard Model Biomedicine came out of stealth with three new papers, each of which illustrates a different combination of the biological scales our multimodal foundation model operates across. One of these papers was titled “Patient-Specific Biomolecular Instruction Tuning of Graph LLMs”. The full text is available on arXiv: https://arxiv.org/pdf/2509.22853.
In this post, we will share further thoughts on this publication and our work behind it.
Biomolecular Instruction Tuning Can Reveal Advanced Molecular Interplays
Omics data is a direct manifestation of biological processes occurring in the human body. While we can measure thousands of biomolecules in cancer patients, understanding what these measurements mean for each individual patient is incredibly complex. These biomolecules (e.g. proteins) aren’t produced in isolation; the measurements that we see are the result of hundreds to thousands of interactions triggering cascades of biological activity.
For example, the p53 tumor suppressor protein orchestrates DNA damage response by activating proteins such as MDM2 and p21 to coordinate cell cycle arrest and DNA repair, preventing the spread of genomic damage. Traditional analysis methods (e.g. univariate t-tests, fold-change ranking, principal component analysis, k-means clustering, logistic regression) analyze these proteomics levels independently, missing this crucial molecular interplay. Even a standard neural network would struggle to learn this distinction, as it must discover from scratch that these specific proteins should be analyzed as a connected group rather than independent features.
This is where biomolecular instruction tuning becomes imperative for analyzing diseases in the human body.
By teaching AI models to understand the patient-specific protein levels within the context of their biomolecular interaction networks, we can capture a more complete biological story. At Standard Model Biomedicine, we expanded upon this idea and developed KRONOS: Knowledge Representation of patient Omics Networks in Oncology via Structured tuning. KRONOS creates an individualized molecular map for every patient by integrating their proteomics data with biomolecular networks, capturing both protein levels and their interaction dynamics within the human body. KRONOS then merges this enriched proteomics representation into an LLM, creating a multi-modal language model that empowers physicians to deliver further personalized care by mapping distinct protein signatures that characterize each patient’s malignancy.
By integrating representations grounded with biological relevance into an LLM, the model captures the complex biology underlying omics measurements, moving beyond pattern recognition towards intricate understanding of molecular networks, dependencies, and cascading effects that drive oncogenesis.
Biomolecular Graphs Boost Deep Learning of Patient Data
While traditional deep learning approaches in biomedicine assume feature independence, integrating biomolecular networks such as STRING PPI enables denoising of multi-omics data and learning of meaningful signals by utilizing these networks as structural priors. Early work such as EMOGI used graph neural networks (GNNs), leveraging these interaction maps to predict cancer genes, while subsequent methods like GNN-SubNet [1] harnessed explainable AI to identify disease-specific subnetworks across a patient cohort. Recent approaches adopt larger and more heterogeneous biomolecular interaction networks (TREE [2]) to better capture these omics complexes. Biomedical AI literature demonstrates that leveraging these molecular networks alongside omics improves learning of meaningful signals in prediction and biological interpretability of diseases.
Alongside these advances in graph omics learning, AI research is adopting new paradigms for enhancing reasoning in LLMs. The introduction of instruction tuning has enabled AI interpretation of modalities in biology and medicine that LLMs may not be already accustomed to.
By training LLMs on medical instructions, researchers enable AI to reason across an out-of-scope modality for complex tasks. MIMIC-Instr [3] allowed LLMs to reason about intricate longitudinal EHR data, while LLaVA-Med [4] learned to expertly understand medical images. MEIT [5] decoded ECG signals into clinical insights, and Me-LLaMA ingested 129 billion biomedical tokens to master medical language. These aren’t just pattern-matching tools anymore, they were AI systems that could understand context, follow clinical reasoning, and translate between different biomedical modalities.
KRONOS: Knowledge Representation of Patient Omics Networks in Oncology via Structured Tuning
While proteomics is imperative to understanding a patient’s disease progression, LLMs haven’t inherently learned how to navigate through this complex modality, let alone how to reason and generate prognostic predictions for a patient’s disease state.
As mentioned above, our paper introduces KRONOS as a framework that integrates enriched patient-specific network representations with modern LLM training paradigms, enabling models to capture complex proteomics signals and achieve competitive prognostic performance.
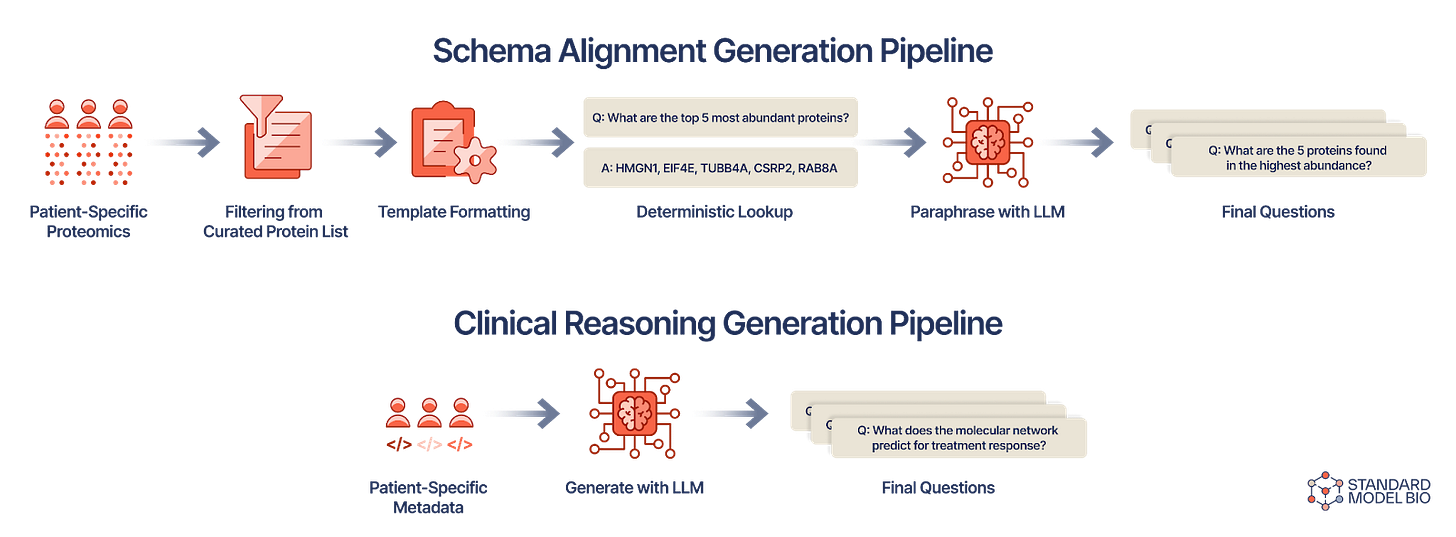
KRONOS grounds patient-specific proteomics representation learning in biomedical interaction networks, generating biologically-relevant embeddings ingested by the LLM. To teach the model this unfamiliar modality, we constructed CPTAC-PROTSTRUCT, a dataset curated from the largest U.S. proteomics cancer study (CPTAC), which forces the LLM to align proteomics signals with its language system and unlocks true reasoning capability. The steps to generate the CPTAC-PROTSTRUCT schema alignment and clinical reasoning instruction pair sets are outlined in the figure above.
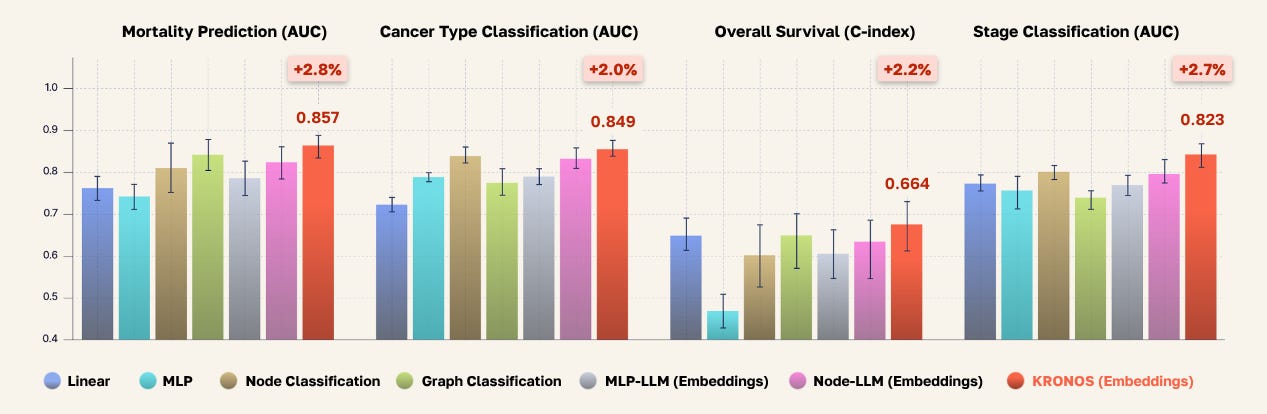
By aligning proteomics with the LLM and enabling reasoning through instruction tuning, the model learns the semantic nuances of this newly-introduced modality. It can then generate proteomics-grounded text and, more importantly, develop rich internal representations that serve as a foundation for various downstream tasks. Once we train KRONOS, we validate the learned representations by probing its hidden layers against ground truth, using a frozen LLM backbone that was not explicitly trained on these downstream tasks during training, shown below.
For comparison, we benchmarked KRONOS against classical and deep learning methods in biomedical literature for predicting prognostic outcomes from omics data. These models range from traditional MLPs, to graph classification of proteomics-injected PPI networks, to multi-modal LLMs with various encoder backbones. Unlike methods specifically trained to predict prognostic outcomes from proteomics data, KRONOS learns general-purpose representations through multi-modal semantic alignment, without task-specific fine-tuning.
Where We Go From Here
KRONOS represents more than just another biomedical AI tool—it’s proof that we can teach language models to fluently speak the language of molecular biology.
Our results show something remarkable: when you combine patient-specific protein networks with instruction-tuned LLMs, you don’t just get better predictions, you get a system that truly understands the biological story behind each patient’s data. The model’s ability to outperform traditional approaches across mortality prediction, cancer typing, and survival analysis demonstrates exciting progress towards even more patient-centric care.
The most exciting implication of our paper is that KRONOS demonstrates the gap between molecular biology and clinical medicine can be bridged by artificial intelligence. This isn’t about replacing doctors or biologists; it’s about giving them a tool that can instantly clinically contextualize complex molecular measurements. As we refine this technology and expand to more biologically-relevant modalities, we move toward a future where treatments are guided by deeper understanding of individual patient molecular landscapes.
For deep dives into our other recent publications and to receive future news and updates from Standard Model Biomedicine, subscribe to our Substack, follow us on X and LinkedIn, or check out our website at www.standardmodel.bio.